Welcome
This introduction help to use the GlycoSim.
Introduction
GlycoSim homepage has mainly 6 tabs
- Glycan Pathway Prediction
- Parameter Setting
- SBML Model Information
- Simulation
- Parameter Estimation
- Sensitivity Analysis
Glycan Pathway Prediction(GPP)
GPP method can predict a glycosylation pathway by using four input information.
Enzyme-Substrate Specificity
Enzyme-substrate specificity defines the reaction relationships between substrates and products. The summary of the information is below:
| Property Name | Introduction | Input rules |
|---|---|---|
| Enzyme | Enzyme name that activates reactions. | Each 'Enzyme' names have to be unique. |
| Substrate | Reactants for reactions. | Its value has to be written in LiCoRR format for describe
substrate-product relationships. Multiple substrates cannot be registered separately with half-spaces. However, They are alternatively registered as one substrates with an under bar. |
| Product | Products for reactions | Same as the 'Substrate' section. |
| Constraint | Products for reactions | Same as the 'Substrate' section. |
| Cosubstrate | Products for reactions | Same as the 'Substrate' section. |
| Class | Enzyme function class that describes whether the enzymes works as a glycosidase or a transferase. | The letters 'GH' or 'GT' has to be specified. 'GH' means a glycosidase, and 'GT' is a transferase. |
| Type | Glycan type of the reactions that describes whether the glycan related to the reaction is a N-glycan, an O-glycan,a L-glycan(Lipid) or an other type of glycan. |
|
| Reaction type | Enzyatic reaction rates |
|
Substrate : (GN$b
Product : (Ab4GN$b
Constraint : none
Cosubstrate : UDP_Gal
Class : GT
Type : O
ReactionType: Sequential-order Bi-Bi kinetics
/
For example, one of the format '$' is used for indicating the modification uncertainty of monosaccharide such as sulfate modification like GN[6S]b3Ab4 wriiten in the LinearCode format. The left parency express the terminal glycan, therefore, the result comes different when the 'Substrate' value is attached with or without a left parency.
The 'Product' property is related to reaction products e.g.) (Ab4GN$b, (...(GNb6)ANa. the product value expression is the LiCoRR format that is same as 'Substrate' value.
The 'Constraint' property attaches substrate conditions to the 'Substrate' value. When these conditions are accomplished, the reaction relationship is attached to the prediction pathway.
The 'Cosubstrate' property defines a co-substrate such as UDP-Gal and UDP-Man. These value have to include hyphen or underbar between words e.g.) UDP-Gal or UDP_Gal for detecting coproduct information.
The 'Class' property can take two enzyme class. The "GH" means a glycosylation hydrase, the "GT" means a glycosylation transferase.
The 'Type' property defines a glycan type. There can be 4 types, N, O, L, and Other. 'N' is stand for N-glycan, 'O' is stand for O-glycan, 'L' is stand for L-glycan.
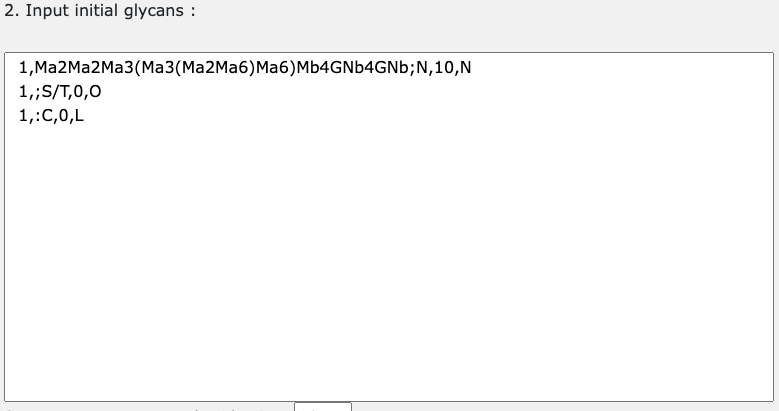
Initial Glycans
 There are four radio buttons for selecting the GPP method.
There are four radio buttons for selecting the GPP method.
The initial glycan is the first substrate for glycan pathway. The input should be 4 comma separated data e.g.) 1,Ma2Ma2Ma3(Ma3(Ma2Ma6)Ma6)Mb4GNb4GNb;N,10,N. 1st tab is the species number, 2nd tab is LinearCode glycan representation, 3rd tab is the number of monosaccharides, 4th is glycan type.
Max monosaccharide
This information is used for decrease the size of the prediction network.
(OPTIONAL) glycoprofile
These optional value is required for 3 prediction method ('forward prediction with glycoprofile', 'backward prediction with initial glycan', 'backward prediction with initial glycan')(
Prediction method
There are 4 types of prediction methods, and the defalt method is forward prediction. This method predicts relationships from intial glycans.
 There are four radio buttons for selecting the GPP method.
There are four radio buttons for selecting the GPP method.
| Method | Summary |
|---|---|
| Forward Prediction | Predict the pathway from the initial glycans. |
| Forward Prediction with glycoprofile | Predict the pathway from the initial glycans, then the size of predicted network is reduced by pruning the unnecessary reactions. |
| Backward Prediction | Predict the pathway retrogradely from the glycoprofile. |
| Backward Prediction with glycoprofile | Predict the pathway retrogradely from the glycoprofile, then the size of predicted network is reduced by pruning the unnecessary reactions. |
Another method, 'forward prediction with glycoprofile' need a glycoprofile information to use network pruning. Backward prediction makes pathway from product glycans. Another method 'Backward prediction with initial glycans' is for network pruning.
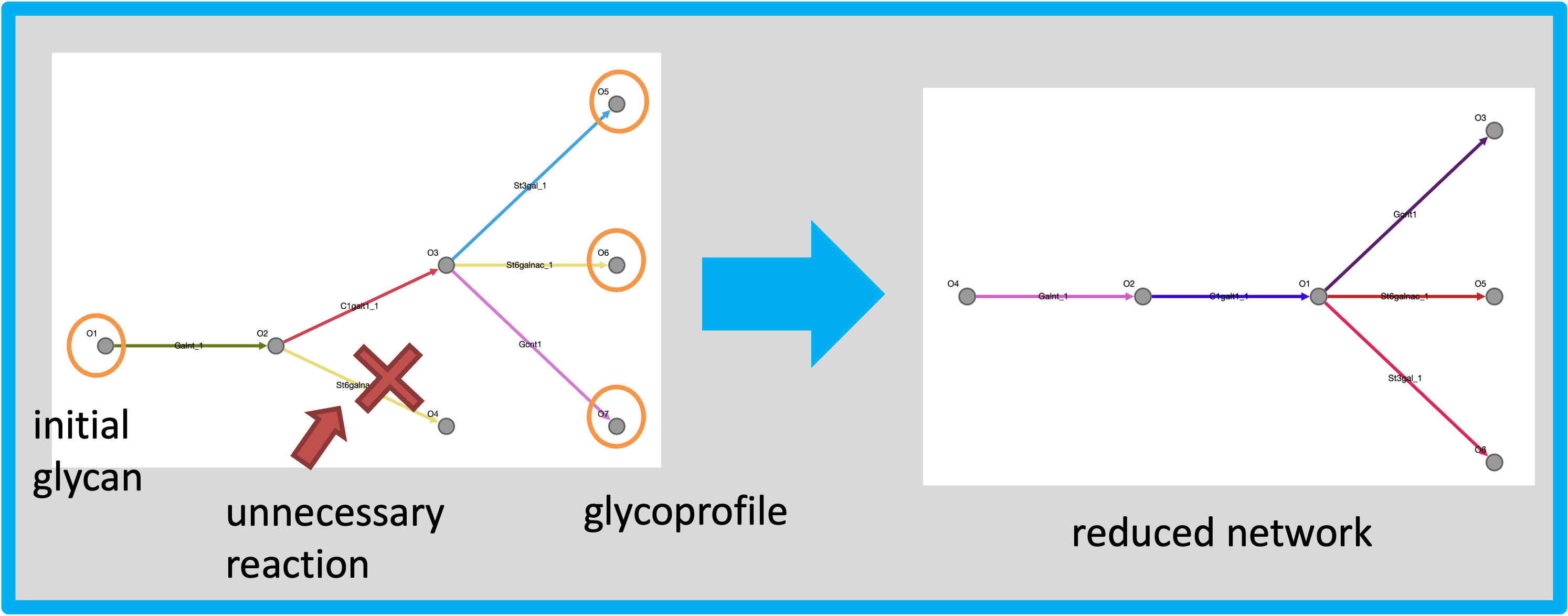 The mechanism of network pruning by the forward prediction with glycoprofile.
After building the reaction network, the glycoprofile are mapped into the predicted network.
Then, we find the pathway retrogradely from the mapped glycans to initial glycans.
The mechanism of network pruning by the forward prediction with glycoprofile.
After building the reaction network, the glycoprofile are mapped into the predicted network.
Then, we find the pathway retrogradely from the mapped glycans to initial glycans.
Result Information
The result for the GPP method gives 3 types of information and prepare data for the next parameter setting section.
- Species information
- Reaction information
- Pathway information
-
Species Information
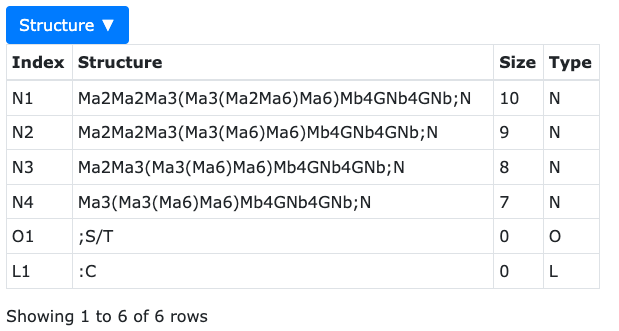 Species information contains 4 columns.
Species information contains 4 columns.Name Summary Index the glycan id for the glycan species. Structure the LinearCode glycan representation Size the length of glycan. Type the glycan type. - Index - the glycan id for the glycan species.
- Structure - the LinearCode glycan representation
- Size - the length of glycan.
- Type - the glycan type.
-
Reaction Information
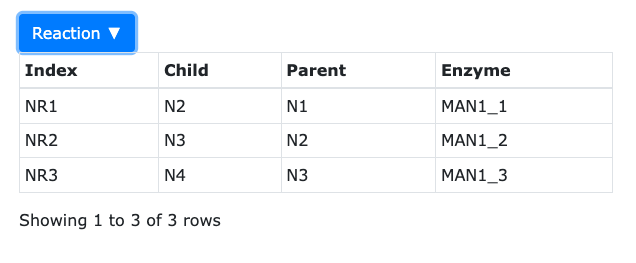 Reactions information contains 4 columns.
Reactions information contains 4 columns.- Index - the reaction id.
- Child - the product species id
- Parent - the reactant species id.
- Enzyme - the enzyme name.
-
Pathway Information
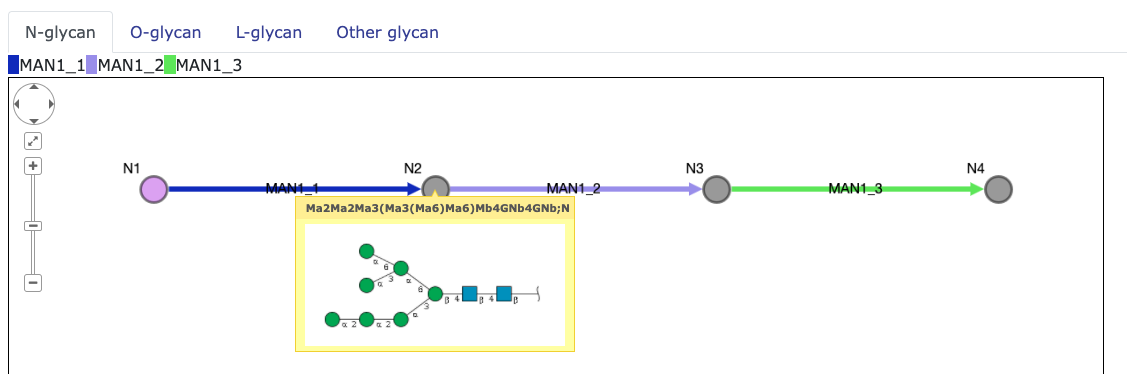 Each node represents the species, and each edges represent the reactions.
When the cursor pointer is on the node, the glycan images are pop up if existed.
Each node represents the species, and each edges represent the reactions.
When the cursor pointer is on the node, the glycan images are pop up if existed.
 There are four radio buttons for selecting the GPP method.
There are four radio buttons for selecting the GPP method.
Parameter Setting
After the pathway prediction, we have to set the parameter sets to build the mathematical model.
Set localization concentration
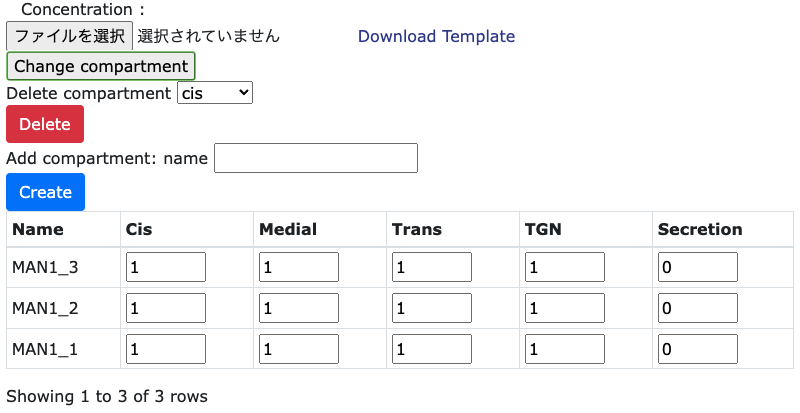 In the left hand side of 'Parameter Setting' page, it is necessary to set the localization values
of each enzymes and sugar nucleotide concentration in each compartments. Users can change the values
directly or downloading the CSV file by clicking the 'Download Template' button. After values are
changed, edited values are relected by uploading the file.
In the left hand side of 'Parameter Setting' page, it is necessary to set the localization values
of each enzymes and sugar nucleotide concentration in each compartments. Users can change the values
directly or downloading the CSV file by clicking the 'Download Template' button. After values are
changed, edited values are relected by uploading the file.
Set kinetic parameters
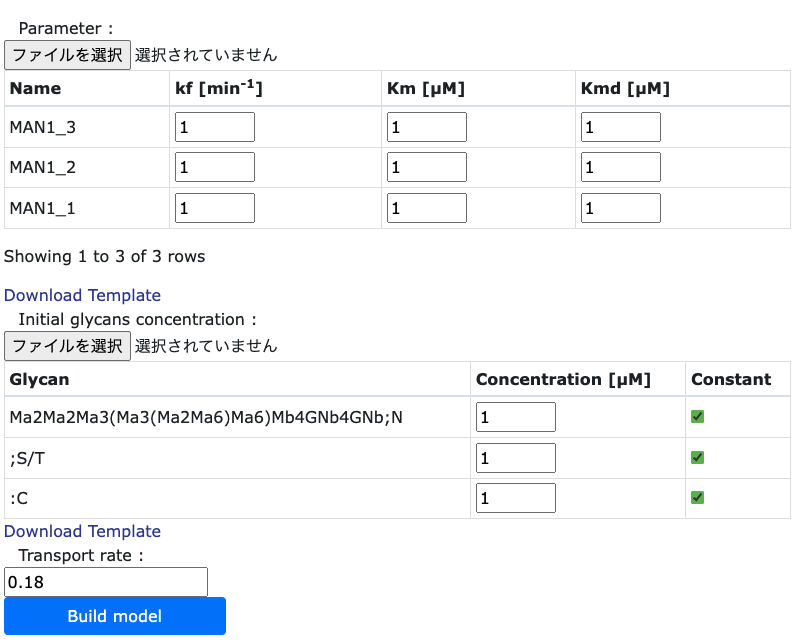 In the right hand side of 'Parameter Setting' page, it is necessary to set the kinetic parameter values
of each enzymes in each compartments. Users can change the values
directly or downloading the CSV file by clicking the 'Download Template' button. This is same as
setting the localization values section.
Note that the kinetic parameters of each reactions shown in the bottom table are changed
when users change the kinetic valus in this table.
In the right hand side of 'Parameter Setting' page, it is necessary to set the kinetic parameter values
of each enzymes in each compartments. Users can change the values
directly or downloading the CSV file by clicking the 'Download Template' button. This is same as
setting the localization values section.
Note that the kinetic parameters of each reactions shown in the bottom table are changed
when users change the kinetic valus in this table.
After that, initial glycans concentration are required. These values can set the value constant or not. Then, transport rates are set, which the defalt value is 0.18 [1/min], which is based on the article.
Set kinetic parameters in each reactions
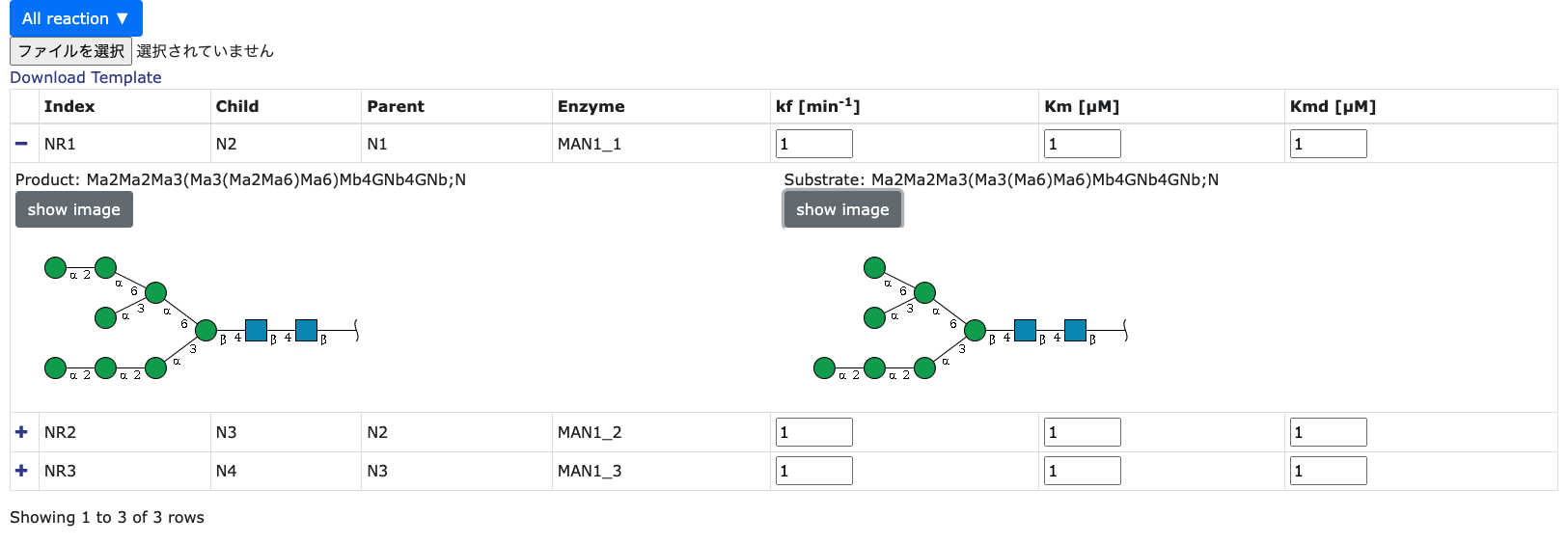 In the bottom side of 'Parameter Setting' page, users can change the kinetic values
of each reactions directly or downloading the CSV file by clicking the 'Download Template' button. This is same as
setting the localization values section.
In the bottom side of 'Parameter Setting' page, users can change the kinetic values
of each reactions directly or downloading the CSV file by clicking the 'Download Template' button. This is same as
setting the localization values section.
Mathematical modeling of golgi apparatus
In the GlycoSim, the concept of the glycosylation model is shown below:
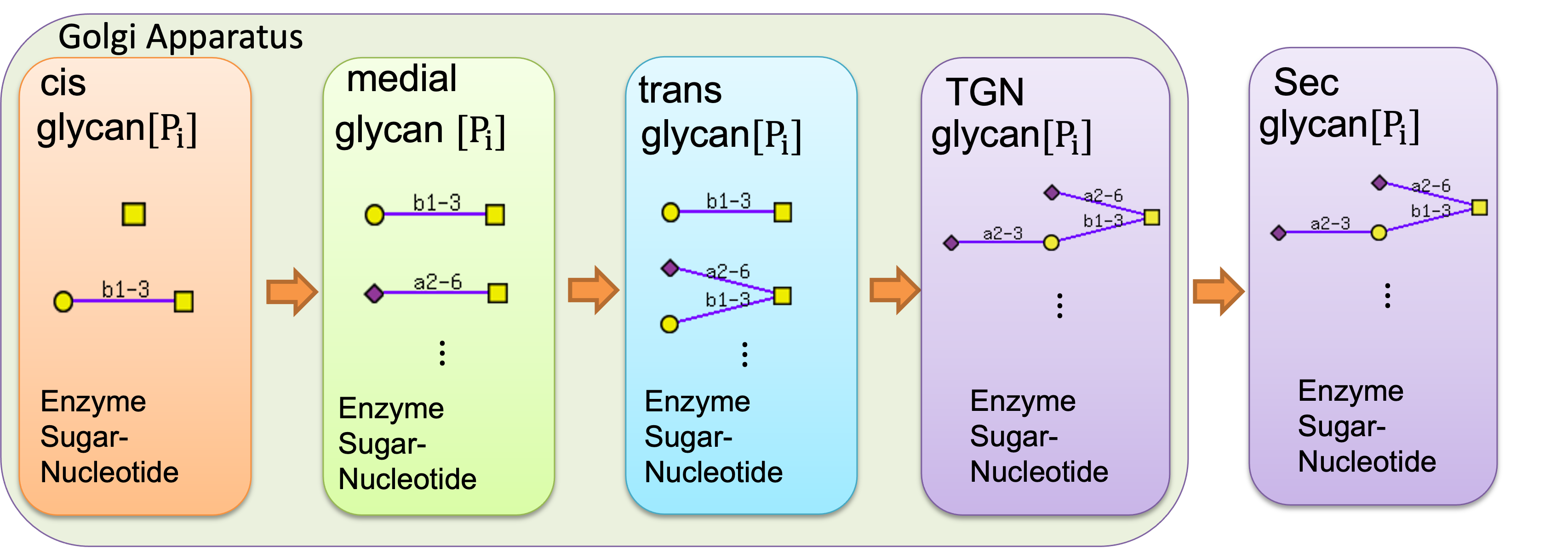 Five compartment model which four of them (cis, medial, trans, TGN) represents a golgi apparatus and the last represents the secretion compartment.
Each compartment can
contains its own glycan species, enzyme species, sugar nucleotide species.
In this model, the glycan species are regarded as an reactants for reactions,
and these species can have different reactions and tranlocations from former compartment
to the latter compartment. Finally, these glycans are translocated to Sec compartment
which represents the compartment apart from golgi compartment.
Five compartment model which four of them (cis, medial, trans, TGN) represents a golgi apparatus and the last represents the secretion compartment.
Each compartment can
contains its own glycan species, enzyme species, sugar nucleotide species.
In this model, the glycan species are regarded as an reactants for reactions,
and these species can have different reactions and tranlocations from former compartment
to the latter compartment. Finally, these glycans are translocated to Sec compartment
which represents the compartment apart from golgi compartment.
Enzyme kinetics
Each reactions in the compartments compose the Michaelis-Menten equation. The example formula of the transferase reactions is below:
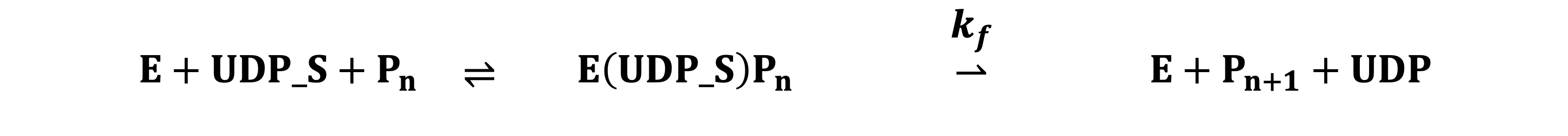
Based on the model, The ordinary differential equations (ODE) are built to simulate the concentration of each glycan species at arbitary timepoints. The example of mathematical formula of the glycosyl transferase is below:
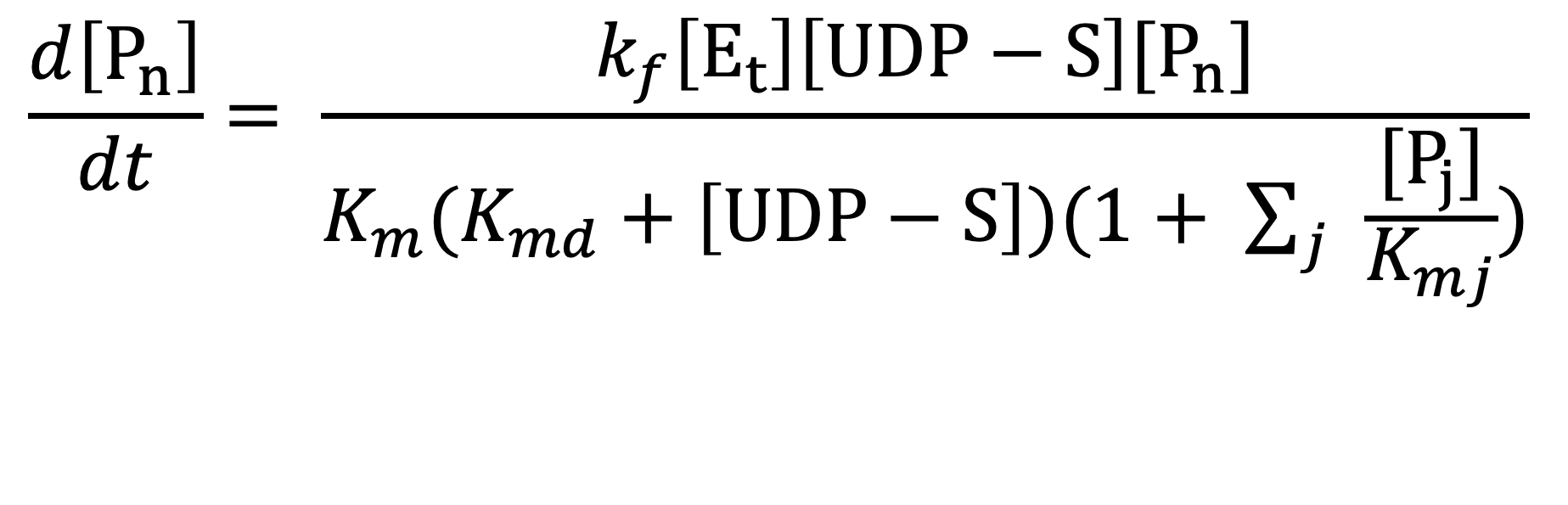
SBML Model Information
'SBML Model Information' page describes the information of the built model. The main component of the SBML file is
- Compartments
- Species
- Parameters
- Reactions
- AssignmentRules
Editing SBML information by double click
Simulation
'Simulation' page describes the information of the built model. The main component of the SBML file is
- SBML information
- Setting duration and time steps
- Solver configuration
- Individual Prot
- Group Plot
- Filtering Plot
Parameter Estimation
'Parameter Estimation' page describes the information of estimation setting, and have 3 pages for adjusting parameter setting, experimental data preparation and result page.
Adjusting parameter page require the estimation parameters by checking the checkbox in the left first column. The number of these parameters should be more than 1 to execute the task. Upper bounds and lower bounds are required to clip the boundary of these values.
After setting the values, click the next 'Experimental Data' tab to translocate the page. In the experimental data table, the left column shows the species names. To set the experimental values, we have to set the experimental time to the first row. Then, we set each species concentration values to each rows. Note that these values should be the same column with 'time' values.
Sensitivity Analysis
'Sensitivity Analysis' page describes the information of Sensitivity setting. In this page, the heatmap of how much the target variables are changed when the effect variables are changed at 1.01% are shown.
Users can select the types of effect variables:
- Parameter
- Reaction
- Initial Condition
- Integral
- Maximum
- Minimum